Review Article | DOI: https://doi.org/10.31579/crr-2022/036
A Study on the Reduction of Airborne Microbial Bioaerosols at Indoor air of Hospital’s Intensive Care Unit by Using Novel Air Filtration and UV Irradiation Technology
- Ioannis Rabias 1*
- Drosos Kourounis 1
- Nikoleta Bobolaki 1
- Nikolaos V. Sipsas 2
- Yanna K. Atanasova 3
- George K. Nicolaides 3
- Maria Evangelidou 4
1 Quality ControlDepartment, Hellenic PasteurInstitute, Athens, Greece.
2 Infectious Diseases Unit, Pathophysiology Department, Laikon General Hospital and National and Kapodistrian University of Athens - Athens, Greece.
3 Department of Mechanical Engineering and Department of Civil Engineering, University of West Attika, Aegaleo,Greece.
4 Diagnostic Department, Hellenic Pasteur Institute, Athens, Greece.
*Corresponding Author: Ioannis Rabias. Quality Control Department, Hellenic Pasteur Institute, Athens, Greece.
Citation: © 2024, Ioannis Rabias, This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Copyright: Ioannis Rabias, Drosos Kourounis, Nikoleta Bobolaki, Maria Evangelidou, Nikolaos V. Sipsas, Ioannis Rabias, (2024). A study on the Reduction of Airborne Microbial Bioaerosols at indoor air of hospital’s Intensive Care Unit by using Novel Air Filtration and UV Irradiation Technology, Clinical Research and Reviews,3(6); DOI:10.31579/crr-2022/036
Received: 18 October 2024 | Accepted: 28 December 2024 | Published: 05 November 2024
Keywords: real time reduction of airborne bioaerosols; air filtration; uvc-ultrapure technology
Abstract
The monitoring of microbial quality and quantity of indoor air in Intensive Care Units is important to make progress at controlling nosocomial infections. This study’s aim was to monitor the presence of pathogenicbacteria and fungi inside an operating Intensive Care Unit and to observe the real-time microbial reduction caused by the application of a prototype air filtration-UV irradiation sterilization device. Air samples of the indoor air of a hospital’s ICU operating room were taken using air sampler technology at opposing sampling spots. The colony formed units were counted, then isolated for the purposes of PCR and molecularsequencing procedures. The microbial burden recovered from the initial sampling was counted at 141 cfu/m3 regarding the bacterial colonies, with species and sub species of Pantoea, Priestia, and Bacillus genre havin a dominant presence. As for the fungi, 112 cfu/m3 were counted, mainly from the Mucor and Aspergillus family. The testing of prototype UVC-ULTRAPURE displayed particularly promising results on the reduction of active airborne microbial particles, with the sterilization success rate beingup to 70%, within 1 hour of the device’sperformance. The need of microbial monitoring in regular basis, inside hospital rooms should be considered, as patients are prone to secondary infections. Future research and surveillance assessing the importance of UVC radiation technology in terminating pathogens.
Introduction
At a worldwidepandemic such as COVID-19, bacteriaand fungi are reported to cause co infections to critically ill patients, which increases morbidity and mortality of the virus [1,2]. Airborne viruses, bacteria, and fungi spores usuallyexist in the form of Bioaerosols. A bio aerosol is an airbornecollection of biological materials. Ubiquitous indoors and out, bio-aerosols in suspended, aerosolized liquid droplets typically contain microbes, like viruses, bacteria and fungi combined with byproducts of cellular metabolism. Inhalation of microbial aerosols can elicitadverse human healtheffects including infection, allergic reaction, inflammation, and respiratory disease [3]. Hospitalization in ICU is highly associated with microbial infections, leading particularly, to ventilator-associated pneumonia (VAP) and bloodstream infections (BSI) [4]. Furthermore, it is observedthat within the first few days after virus infection, critically ill patients often develop respiratory tract distortion or pulmonary dysbiosis, which can furtherprogress into a secondary bacterialor fungal infection just a few weeks later[5]. The rapid and accurate identification of bacteria or fungi, that present as pathogenic or resident microorganisms inside hospital wards, during the period of COVID-19 should be an important step towards the management of patients, especially when there are several published papers [6, 7, 8] which claim, that bacteria and fungi can act as vessels for viruses and by doing so, these microorganisms can demonstrate a coinfection ability (aiming their own proliferation) or can act supportively to virus’s transmission. Another crucial factor for the reduction of the virus’s spread is the de-contamination of the hospital’s Covid-ward indoor air, by means which do not harm the patientand also do not impedethe daily routineof the personnel. In this work we performed a two-scale experimental procedure, which aim was to initially identify the genre of the microbial load inside a hospital’s Intensive Care Unit and furthermore, to investigate the decontamination potential of a prototype air sterilization device,UVC ULTRAPURE, invented at the Department of Mechanical Engineering at the University of West Attica Greece.The device’s operating principle is basedon the fact that pathogens are being captured on the surface of a wide angle (WA) HEPA FILTER, where this new type of HEPA filter has the capacity to restrain 99.7% of pathogens (bacteria, fungi, and virus etc.) due to the 0.1 to 0.3 micrometer (μm) size of WA HEPA filter holes. It is worth to mention that the size of virus particlesis at the scale of 0.1μm, but they are present almostalways within larger droplets (>0.1μm), generated by infected hosts and therefore almost always contained by (WA) HepaFilter. Contrary to a regular HEPA filter which has dense folds, WA HEPA filter has larger angle between foldsto avoid the creation of shading effectsduring its irradiation and therefore its entire surfacecan be continuously UV irradiated. Furthermore, the irradiation is also being reinforced by a quartz grid –opticalgrating which createsa multi – focus effectof UV light on boththe surface and holes of HEPA through an optical multi diffraction effect. The capture of microorganisms and the subsequent UV continuous irradiation results in the total distraction of pathogen’s organicmaterial without leavingbehind any organicresidues on the filter. Consequently, any saturation of the filterwill be due only to inorganic material (i.e., dust), thus eliminating the health risk related with the handlingof filter replacement and disposal. The need for search for a better UV technology air sterilizer aroused after realizing that most of the devices that are available in the market using UV irradiation for pathogen neutralization and destruction are using the technique of irradiating an air stream which passes through the device and contains these pathogens. According though to published results, the dwelt time of pathogens in the irradiated zone for these devices is extremely small,thus the UV radiation doesn’thave the necessary time to kill these pathogens. To the contrary,the proposed new technology is not based on the irradiation of an air stream, but instead on the capturing first of the pathogens on a flat WA HEPA filter surface and subsequently through intense and continuous multi focused irradiation, eliminates them.
Methods
Monitoring of Microbial Load.
The microbial load’sidentification process consistsof several steps, starting with air sampling from the hospital’s ward space, which had a size of approximately 90 m3 and was occupiedby 2 patients at their recovery phase.Through air sampling, it is possible to evaluate microbial contamination in environments with considerable risk of infection. Microbial load measurements with air samplers are performed in accordance with the EU GMP for proper manufacturing principles for microbialmonitoring of ambient air in controlled environments up to clean rooms [9]. The site’s microbial load was measured by using MAS-100 air sampler (Figure 1), which consists of a radial fan, controlled by a flow sensor, that accurately regulates the real time air flow at 100 liters/min for a 10 min time interval. A specified amount of air is aspirated through a perforated lid and impacted onto the surface of growth media inside a 90-100 mm Petri dish. This procedure is called ‘active air monitoring’, in contrast with ‘passive air monitoring’, where the sampling is performed throughgravity force only and is a more time-consuming technique[10].

Figure 1: The MAS-100 air samplerand Petri dishes positioning on the room and sampling.
Two types of Petri dishes are used (Figure.1), one that containsTSA (Tryptone soya agar) growth medium for bacteria promotion and another with SAB (Sabouraud agar) medium for fungi promotion. After the standardPetri dish’s incubation time (according to ISO 14698-1 instructions), the results found regarding the number of bacteria or fungi, are expressed in cfu (colony forming units) / m3. For a better estimation of the microbial load inside the ICU, air samples were taken in two distinct spots, the first one locating between the patient’s head high and the second one at the room’s center (Figure 2).
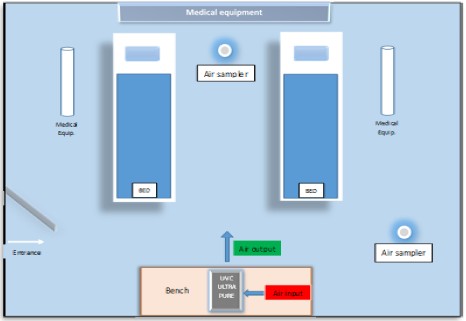
Figure 2: Graphical representation of the Intensive Care Unit (ICU). The air sterilizing device (silver) filtrates air input (red) through WA HEPA, radiates it with UVC and air output (green) is released back in the room.
All doors and windows were kept locked during the sampling, in order to keep theexperiment conditions unaltered from exterior factors Decontamination Potential of a Prototype Air Sterilization Device UVC ULTRAPURE, as the device is called, can filtrate air and apply UVC radiation using a novel patterned technique. The air passes through HEPA 13 filters, which have the ability of retaining microbial particles [13]. These particles are subsequently UVC irradiated, a procedure that can kill most ofthe retained microbes [14] and moreover, the emitted radiation does not encounterindividuals around it, because of the device’ssteel plated protection. Herein we evaluated the performance of UVC ULTRAPURE prototype, which apply cutting edge technologies, mentioned in the above references. The second scale of this work was to examine the sterilization potential of UVC ULTRAPURE, inside a rather contaminated environment, as an ICU theoretically is and at real life conditions (patients inside experimental area) betweentwo different time points, 30 and 60 min after the device’sapplication. These 30-minute interval between measurements was decided upon the fact that each measurement takes approximately 10 minutes to complete. As in the first sampling, two distinct sampling spots were used, specimens for TSA and Sabouraud plates were taken and also all doors and windowswere kept shut.Following the samplingprocedure, the plates were incubated at the appropriate conditions (32.5 °C for TSA plates and 22.5 °C for SAB).
Molecular Identification
The last step of the experiment’s procedurewas to identify the genre of thesemicrobial colonies and by doing so to have an estimation of the microorganism’s pathogenicity, which ifexisting, has a crucial effecton the patient’s health and recovery. To achieve that, single colonies from agar plates were isolated and diluted in 2ml PBS, so that to be processed for DNA extraction using the Mag Core automated nucleic acid extractor, according to the manufacturer’s protocol. DNA samples extracted from the TSA and Sabouraud agar plates were subjected to Polymerase Chain Reaction using primers targetingthe 16S ribosomal RNA and 18S ribosomalRNA respectively. PCR products were analyzed using the QIAxcel Advanced System. All three DNA samples extracted from TSA agar plates colonies were found positive for 16S ribosomal RNA and 2 out of 5 DNA samples extracted from Sabouraud agar plates colonies were found positive for 18S ribosomal RNA. Once validated, the PCR products were purified using the QIAquick PCR purification kit according to the manufacturer’s protocol. DNA concentration was measured using the Nanodrop spectrophotometer. Sanger sequencing was performed in 120-150 ngr/ reaction of DNA using the internal primers 536F, 536R, 800F, 800R, 1050F and 1050R for 16S ribosomal RNA as previously described and the NS5F, NS6R for the 18S ribosomal RNA. Sequencing data were processedusing the BioEditSoftware and consensussequences were analyzedby BLAST, with over 90% successful identification rate for each of the isolated microbial colonies that were recovered.
Results
At the end of incubation period the number of microorganisms before and after the application of the prototype device was recorded and also the percentage for the reductionof microorganisms due to the use of the sterilization system,was measured. From the mean values of sampling, came the followingresults of air samplesmeasurements (Figure.3).
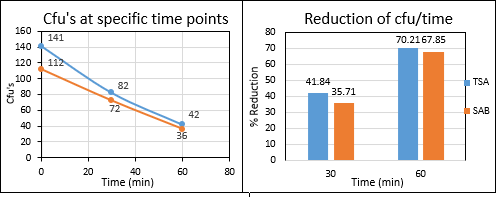
Figure 3: Representation of the reduction of counted colonies and of ?u’s reduction as sterilization time proceeds (cfu/time) by air samples Measurements of bacterial and fungal coloniescounted at standardtime points beforeand after device’sapplication.
Specifically, it was found that the device contributed to a 70.21% reduction in bacterial load, as well as a 67.85%reduction in fungalload. At this point, it is worth mentioning that our initial measurements (before the application of the sterilization system)present to us a spacewith a fairly heavy microbial load, as for both bacteria and fungi more than 100 cfu / plate were retrieved for 1 m3 of air sample. After 1 hour of the device’s application and with 4 people always present inside the room (2 patients and 2 research analysts) one can observe that the sterilization system proceeds to a steadilyreduction of the air’s microbial load. According to the number of cfu/m3 that were recovered from the sampling procedure, it is safe to claim that the ICU’s microbial load was relatively high, in relation to clean room standards for GMP (Good Manufacturing Practice), which is justified when patient’s contamination biomaterial is constantly spreading inside the room. Τhe results from BLAST sequencing which came out, showedthat the recoveredcolonies from samplingbelonged to the following genera-species of bacteria and fungi(Table 1 and Table 2).
Bacterial colonies | |
|
|
|
|
|
|
|
|
|
|
| |
Table 1: Genera-Species of Bacteria from the recovered Coloniesof Sampling.
The habitat of the recoveredbacterial species is placed either in the microfauna of the human digestive system, or in the external environment (comingfrom the soil) or foundin certain foods (such as vegetables). Due to the nature of the experimental field (ICU unit), it was considered necessary to examine the pathogenicity of the identified microorganisms in the human immunological system. As previously mentioned, among the bacterial genera recovered there were several species-representatives of the genus Pantoea, which [11] are potentially pathogenic in immunocompromised patients.Examining the rest of the bacterial genera(Priestia, Neobacillus and Bacillus) no specific reference to the existence of pathogenicity was found, but this does not exclude the occurrence of pathogenicity in case of an overpopulation of thesaid bacterial genera.
Fungal colonies | |
|
|
|
|
|
|
|
brasilliensis) |
Table 2: genera-species of fungi from the recovered colonies of sampling.
The fungal genera-species that were recovered are all microorganisms of wide and even global distribution, coming mainly from food, from the microbiome of the humanskin, or from the externalenvironment. Regarding theirpotential pathogenicity, the genus Aspergillus [12], in specificconditions of aspiration of large amountsof spores of the fungus,can lead to the appearance of aspergillosis, a serious disease of the respiratory system. The species of the genus Mucor do not show pathogenicity in humans, due to their lack of ability to reproduce at 36-37 ºC, apart from some thermo- resistantstrains, which were not found in the fungi we examined.
Discussion
Admittedly, this study faced limitations regarding the repeatability of results, which can be justified due to bureaucratic issues that made it difficult torepeat the experiment. The aim of this study, as mentioned above, was to initiallyperform a monitoring of microbiological materialinside an ICU and furthermore to assess the sterilization potentialof UVC ULTRAPURE device. As for the first step, the results from the air sampling indicate that insidethe ICU the microbial load was relatively high, something that can be justified from the existenceof 2 patients inside the room. Althoughthe hospital’s personnel made efforts to keep the environment clean with daily cleaning procedures, the continuous emission of biological material from the patients and the importation of microbes from external environment, kept the bioburden at high values.According to the molecular sequencing of colonies recovered from TSA and SAB plates,a range of potential pathogenic bacteria and fungi was found,which can (underspecific circumstances) have a severeimpact on the immune systemof already immunosuppressed patients. Those findings strengthen the opinion that the conventional means of cleaning are insufficient and extra sterilizing of the ICU is indeed needed.
Conclusion
The prototype UVC ULTRAPURE sterilization device had undergonebefore similar testing inside areas of public interest (classroom, bus, metro) with worth mentioning results, achieving microbial reduction at a range of 45 up to 93% within 30 minutes of system’s application [15]. In this work the device was evaluated inside a specific nosocomial area (ICU) with continuous feedback of biological matter coming from the patients and the results were once more encouraging with an estimated reduction up to 70% within 60 minutes of usage. These results imply that, despitethe presence of patients and the personnel conducting the experiment, the device has a sterilization potency which is not hindered by the emission of an amount of biological matter inside the experimental zone.
Author Contributions:
All authors have contributed to this work equally, both in the execution of the experiment and in the writing process.
Acknowledgements:
Thankingfor the official cooperation Mr. Fanis Roides,President of Administration Board of the General University Hospital "LAIKO"-Athens Greece.
Conflicts of Interest:
“The authors declareno conflict of interest.
References
- Heidari (2017). “Different High–Resolution Simulations of Medical, Medicinal, Clinical, Pharmaceutical and Therapeutics Oncology of Human Lung Cancer Translational Anti–Cancer Nano Drugs Delivery Treatment Process under Synchrotron and X–Ray Radiations”, J Med Oncol. Vol. 1 No. 1: 1,
View at Publisher | View at Google Scholar - Heidari, “A Modern Ethnomedicinal Technique for Transformation, Prevention and Treatment of Human Malignant Gliomas Tumors into Human Benign Gliomas Tumors under Synchrotron Radiation”, Am J Ethnomed, Vol. 4 No. 1: 10.
View at Publisher | View at Google Scholar - Heidari, (2017). “Active Targeted Nanoparticles for Anti–Cancer Nano Drugs Delivery across the Blood–Brain Barrier for Human Brain Cancer Treatment, Multiple Sclerosis (MS) and Alzheimer's Diseases Using Chemical Modifications of Anti–Cancer Nano Drugs or Drug–Nanoparticles through Zika Virus (ZIKV) Nanocarriers under Synchrotron Radiation”, J Med Chem Toxicol, 2 (3): 1–5.
View at Publisher | View at Google Scholar - Heidari, (2017). “Investigation of Medical, Medicinal, Clinical and Pharmaceutical Applications of Estradiol, Mestranol (Norlutin), Norethindrone (NET), Norethisterone Acetate (NETA), Norethisterone Enanthate (NETE) and Testosterone Nanoparticles as Biological Imaging, Cell Labeling, Anti–Microbial Agents and Anti–Cancer Nano Drugs in Nanomedicines Based Drug Delivery Systems for Anti–Cancer Targeting and Treatment”, Parana Journal of Science and Education (PJSE)–v.3, n.4, (10–19) October 12.
View at Publisher | View at Google Scholar - Heidari, (2017). “A Comparative Computational and Experimental Study on Different Vibrational Biospectroscopy Methods, Techniques and Applications for Human Cancer Cells in Tumor Tissues Simulation, Modeling, Research, Diagnosis and Treatment”, Open J Anal Bioanal Chem 1 (1): 014–020.
View at Publisher | View at Google Scholar - Heidari, (2017). “Combination of DNA/RNA Ligands and Linear/Non–Linear Visible–Synchrotron Radiation–Driven N–Doped Ordered Mesoporous Cadmium Oxide (CdO) Nanoparticles Photocatalysts Channels Resulted in an Interesting Synergistic Effect Enhancing Catalytic Anti–Cancer Activity”, Enz Eng 6: 1.
View at Publisher | View at Google Scholar - Heidari, (2017). “Modern Approaches in Designing Ferritin, Ferritin Light Chain, Transferrin, Beta–2 Transferrin and Bacterioferritin–Based Anti–Cancer Nano Drugs Encapsulating Nanosphere as DNA–Binding Proteins from Starved Cells (DPS)”, Mod Appro Drug Des. 1 (1). MADD.000504.
View at Publisher | View at Google Scholar - Heidari, (2017). “Potency of Human Interferon β–1a and Human Interferon β–1b in Enzymotherapy, Immunotherapy, Chemotherapy, Radiotherapy, Hormone Therapy and Targeted Therapy of Encephalomyelitis Disseminate/Multiple Sclerosis (MS) and Hepatitis A, B, C, D, E, F and G Virus Enter and Targets Liver Cells”, J Proteomics Enzymol 6: 1.
View at Publisher | View at Google Scholar - Heidari, (2017). “Transport Therapeutic Active Targeting of Human Brain Tumors Enable Anti–Cancer Nanodrugs Delivery across the Blood–Brain Barrier (BBB) to Treat Brain Diseases Using Nanoparticles and Nanocarriers under Synchrotron Radiation”, J Pharm Pharmaceutics 4 (2): 1–5.
View at Publisher | View at Google Scholar - Heidari, (2017). C. Brown, “Combinatorial Therapeutic Approaches to DNA/RNA and Benzylpenicillin (Penicillin G), Fluoxetine Hydrochloride (Prozac and Sarafem), Propofol (Diprivan), Acetylsalicylic Acid (ASA) (Aspirin), Naproxen Sodium (Aleve and Naprosyn) and Dextromethamphetamine Nanocapsules with Surface Conjugated DNA/RNA to Targeted Nano Drugs for Enhanced Anti–Cancer Efficacy and Targeted Cancer Therapy Using Nano Drugs Delivery Systems”, Ann Adv Chem. 1 (2): 061–069.
View at Publisher | View at Google Scholar - Heidari, (2017). “High–Resolution Simulations of Human Brain Cancer Translational Nano Drugs Delivery Treatment Process under Synchrotron Radiation”, J Transl Res. 1 (1): 1–3.
View at Publisher | View at Google Scholar - Heidari, (2017). “Investigation of Anti–Cancer Nano Drugs’ Effects’ Trend on Human Pancreas Cancer Cells and Tissues Prevention, Diagnosis and Treatment Process under Synchrotron and X–Ray Radiations with the Passage of Time Using Mathematica”, Current Trends Anal Bioanal Chem, 1 (1): 36–41.
View at Publisher | View at Google Scholar - Heidari, (2017). “Pros and Cons Controversy on Molecular Imaging and Dynamics of Double–Standard DNA/RNA of Human Preserving Stem Cells–Binding Nano Molecules with Androgens/Anabolic Steroids (AAS) or Testosterone Derivatives through Tracking of Helium–4 Nucleus (Alpha Particle) Using Synchrotron Radiation”, Arch Biotechnol Biomed. 1 (1): 067–0100.
View at Publisher | View at Google Scholar - Heidari, (2017). “Visualizing Metabolic Changes in Probing Human Cancer Cells and Tissues Metabolism Using Vivo 1H or Proton NMR, 13C NMR, 15N NMR and 31P NMR Spectroscopy and Self–Organizing Maps under Synchrotron Radiation”, SOJ Mater Sci Eng 5 (2): 1–6.
View at Publisher | View at Google Scholar - Heidari, (2017). “Cavity Ring–Down Spectroscopy (CRDS), Circular Dichroism Spectroscopy, Cold Vapour Atomic Fluorescence Spectroscopy and Correlation Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Enliven: Challenges Cancer Detect Ther 4 (2): e001.
View at Publisher | View at Google Scholar - Heidari, (2017). “Laser Spectroscopy, Laser–Induced Breakdown Spectroscopy and Laser–Induced Plasma Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Int J Hepatol Gastroenterol, 3 (4): 079–084.
View at Publisher | View at Google Scholar - Heidari, (2017). “Time–Resolved Spectroscopy and Time–Stretch Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Enliven: Pharmacovigilance and Drug Safety 4 (2): e001.
View at Publisher | View at Google Scholar - Heidari, (2017). “Overview of the Role of Vitamins in Reducing Negative Effect of Decapeptyl (Triptorelin Acetate or Pamoate Salts) on Prostate Cancer Cells and Tissues in Prostate Cancer Treatment Process through Transformation of Malignant Prostate Tumors into Benign Prostate Tumors under Synchrotron Radiation”, Open J Anal Bioanal Chem 1 (1): 021–026.
View at Publisher | View at Google Scholar - Heidari, (2017). “Electron Phenomenological Spectroscopy, Electron Paramagnetic Resonance (EPR) Spectroscopy and Electron Spin Resonance (ESR) Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Austin J Anal Pharm Chem. 4 (3): 1091.
View at Publisher | View at Google Scholar - Heidari, (2017). “Therapeutic Nanomedicine Different High–Resolution Experimental Images and Computational Simulations for Human Brain Cancer Cells and Tissues Using Nanocarriers Deliver DNA/RNA to Brain Tumors under Synchrotron Radiation with the Passage of Time Using Mathematica and MATLAB”, Madridge J Nano Tech. Sci. 2 (2): 77–83.
View at Publisher | View at Google Scholar - Heidari, (2017). “A Consensus and Prospective Study on Restoring Cadmium Oxide (CdO) Nanoparticles Sensitivity in Recurrent Ovarian Cancer by Extending the Cadmium Oxide (CdO) Nanoparticles–Free Interval Using Synchrotron Radiation Therapy as Antibody–Drug Conjugate for the Treatment of Limited–Stage Small Cell Diverse Epithelial Cancers”, Cancer Clin Res Rep, 1: 2, e001.
View at Publisher | View at Google Scholar - Heidari, (2017). “A Novel and Modern Experimental Imaging and Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under White Synchrotron Radiation”, Cancer Sci Res Open Access 4 (2): 1–8.
View at Publisher | View at Google Scholar - Heidari, (2017). “Different High–Resolution Simulations of Medical, Medicinal, Clinical, Pharmaceutical and Therapeutics Oncology of Human Breast Cancer Translational Nano Drugs Delivery Treatment Process under Synchrotron and X–Ray Radiations”, J Oral Cancer Res 1 (1): 12–17.
View at Publisher | View at Google Scholar - Heidari, (2017). “Vibrational Decihertz (dHz), Centihertz (cHz), Millihertz (mHz), Microhertz (μHz), Nanohertz (nHz), Picohertz (pHz), Femtohertz (fHz), Attohertz (aHz), Zeptohertz (zHz) and Yoctohertz (yHz) Imaging and Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, International Journal of Biomedicine, 7 (4), 335–340.
View at Publisher | View at Google Scholar - Heidari, (2017). “Force Spectroscopy and Fluorescence Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, EC Cancer, 2 (5), 239–246.
View at Publisher | View at Google Scholar - Heidari, (2017). “Photoacoustic Spectroscopy, Photoemission Spectroscopy and Photothermal Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, BAOJ Cancer Res Ther, 3: 3, 045–052.
View at Publisher | View at Google Scholar - Heidari, (2017). “J–Spectroscopy, Exchange Spectroscopy (EXSY), Nucle¬ar Overhauser Effect Spectroscopy (NOESY) and Total Correlation Spectroscopy (TOCSY) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, EMS Eng Sci J, 1 (2): 006–013.
View at Publisher | View at Google Scholar - Heidari, (2017). “Neutron Spin Echo Spectroscopy and Spin Noise Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Int J Biopharm Sci, 1: 103–107.
View at Publisher | View at Google Scholar - Heidari, (2017). “Vibrational Decahertz (daHz), Hectohertz (hHz), Kilohertz (kHz), Megahertz (MHz), Gigahertz (GHz), Terahertz (THz), Petahertz (PHz), Exahertz (EHz), Zettahertz (ZHz) and Yottahertz (YHz) Imaging and Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Madridge J Anal Sci Instrum, 2 (1): 41–46.
View at Publisher | View at Google Scholar - Heidari, (2018). “Two–Dimensional Infrared Correlation Spectroscopy, Linear Two–Dimensional Infrared Spectroscopy and Non–Linear Two–Dimensional Infrared Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, J Mater Sci Nanotechnol 6 (1): 101.
View at Publisher | View at Google Scholar - Heidari, (2018). “Fourier Transform Infrared (FTIR) Spectroscopy, Near–Infrared Spectroscopy (NIRS) and Mid–Infrared Spectroscopy (MIRS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, Int J Nanotechnol Nanomed, Volume 3, Issue 1, Pages 1–6.
View at Publisher | View at Google Scholar - Heidari, (2018). “Infrared Photo Dissociation Spectroscopy and Infrared Correlation Table Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, Austin Pharmacol Pharm, 3 (1): 1011.
View at Publisher | View at Google Scholar - Heidari, (2018). “Novel and Transcendental Prevention, Diagnosis and Treatment Strategies for Investigation of Interaction among Human Blood Cancer Cells, Tissues, Tumors and Metastases with Synchrotron Radiation under Anti–Cancer Nano Drugs Delivery Efficacy Using MATLAB Modeling and Simulation”, Madridge J Nov Drug Res, 1 (1): 18–24.
View at Publisher | View at Google Scholar - Heidari, (2018). “Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Open Access J Trans Med Res, 2 (1): 00026–00032.
View at Publisher | View at Google Scholar - M. R. R. Gobato, R. Gobato, A. Heidari, (2018). “Planting of Jaboticaba Trees for Landscape Repair of Degraded Area”, Landscape Architecture and Regional Planning, Vol. 3, No. 1, Pages 1–9.
View at Publisher | View at Google Scholar - Heidari, (2018). “Fluorescence Spectroscopy, Phosphorescence Spectroscopy and Luminescence Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, SM J Clin. Med. Imaging, 4 (1): 1018.
View at Publisher | View at Google Scholar - Heidari, (2018). “Nuclear Inelastic Scattering Spectroscopy (NISS) and Nuclear Inelastic Absorption Spectroscopy (NIAS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Int J Pharm Sci, 2 (1): 1–14.
View at Publisher | View at Google Scholar - Heidari, (2018). “X–Ray Diffraction (XRD), Powder X–Ray Diffraction (PXRD) and Energy–Dispersive X–Ray Diffraction (EDXRD) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, J Oncol Res; 2 (1): 1–14.
View at Publisher | View at Google Scholar - Heidari, (2018). “Correlation Two–Dimensional Nuclear Magnetic Reso¬nance (NMR) (2D–NMR) (COSY) Imaging and Spectrosco¬py Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, EMS Can Sci, 1–1–001.
View at Publisher | View at Google Scholar - Heidari, (2018). “Thermal Spectroscopy, Photothermal Spectroscopy, Thermal Microspectroscopy, Photothermal Microspectroscopy, Thermal Macrospectroscopy and Photothermal Macrospectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, SM J Biometrics Biostat, 3 (1): 1024.
View at Publisher | View at Google Scholar - Heidari, (2018). “A Modern and Comprehensive Experimental Biospectroscopic Comparative Study on Human Common Cancers’ Cells, Tissues and Tumors before and after Synchrotron Radiation Therapy”, Open Acc J Oncol Med. 1 (1).
View at Publisher | View at Google Scholar - Heidari, (2018). “Heteronuclear Correlation Experiments Such as Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC), Heteronuclear Multiple–Quantum Correlation Spectroscopy (HMQC) and Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC) Comparative Study on Malignant and Benign Human Endocrinology and Thyroid Cancer Cells and Tissues under Synchrotron Radiation”, J Endocrinol Thyroid Res, 3 (1): 555603.
View at Publisher | View at Google Scholar - Heidari, (2018). “Nuclear Resonance Vibrational Spectroscopy (NRVS), Nuclear Inelastic Scattering Spectroscopy (NISS), Nuclear Inelastic Absorption Spectroscopy (NIAS) and Nuclear Resonant Inelastic X–Ray Scattering Spectroscopy (NRIXSS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Int J Bioorg Chem Mol Biol. 6 (1e): 1–5.
View at Publisher | View at Google Scholar - Heidari, (2018). “A Novel and Modern Experimental Approach to Vibrational Circular Dichroism Spectroscopy and Video Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under White and Monochromatic Synchrotron Radiation”, Glob J Endocrinol Metab. 1 (3). GJEM. 000514–000519.
View at Publisher | View at Google Scholar - Heidari, (2018). “Pros and Cons Controversy on Heteronuclear Correlation Experiments Such as Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC), Heteronuclear Multiple–Quantum Correlation Spectroscopy (HMQC) and Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, EMS Pharma J. 1 (1): 002–008.
View at Publisher | View at Google Scholar - Heidari, (2018). “A Modern Comparative and Comprehensive Experimental Biospectroscopic Study on Different Types of Infrared Spectroscopy of Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, J Analyt Molecul Tech. 3 (1): 8.
View at Publisher | View at Google Scholar - Heidari, (2018). “Investigation of Cancer Types Using Synchrotron Technology for Proton Beam Therapy: An Experimental Biospectroscopic Comparative Study”, European Modern Studies Journal, Vol. 2, No. 1, 13–29.
View at Publisher | View at Google Scholar - Heidari, (2018). “Saturated Spectroscopy and Unsaturated Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Imaging J Clin Medical Sci. 5 (1): 001–007.
View at Publisher | View at Google Scholar - Heidari, (2018). “Small–Angle Neutron Scattering (SANS) and Wide–Angle X–Ray Diffraction (WAXD) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Int J Bioorg Chem Mol Biol. 6 (2e): 1–6.
View at Publisher | View at Google Scholar - Heidari, (2018). “Investigation of Bladder Cancer, Breast Cancer, Colorectal Cancer, Endometrial Cancer, Kidney Cancer, Leukemia, Liver, Lung Cancer, Melanoma, non–Hodgkin Lymphoma, Pancreatic Cancer, Prostate Cancer, Thyroid Cancer and Non–Melanoma Skin Cancer Using Synchrotron Technology for Proton Beam Therapy: An Experimental Biospectroscopic Comparative Study”, Ther Res Skin Dis 1 (1).
View at Publisher | View at Google Scholar - Heidari, (2018). “Attenuated Total Reflectance Fourier Transform Infrared (ATR–FTIR) Spectroscopy, Micro–Attenuated Total Reflectance Fourier Transform Infrared (Micro–ATR–FTIR) Spectroscopy and Macro–Attenuated Total Reflectance Fourier Transform Infrared (Macro–ATR–FTIR) Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, International Journal of Chemistry Papers, 2 (1): 1–12.
View at Publisher | View at Google Scholar - Heidari, (2018). “Mössbauer Spectroscopy, Mössbauer Emission Spectroscopy and 57Fe Mössbauer Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Acta Scientific Cancer Biology 2.3: 17–20.
View at Publisher | View at Google Scholar - Heidari, (2018). “Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, Organic & Medicinal Chem IJ. 6 (1): 555676.
View at Publisher | View at Google Scholar - Heidari, (2018). “Correlation Spectroscopy, Exclusive Correlation Spectroscopy and Total Correlation Spectroscopy Comparative Study on Malignant and Benign Human AIDS–Related Cancers Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Int J Bioanal Biomed. 2 (1): 001–007.
View at Publisher | View at Google Scholar - Heidari, (2018). “Biomedical Instrumentation and Applications of Biospectroscopic Methods and Techniques in Malignant and Benign Human Cancer Cells and Tissues Studies under Synchrotron Radiation and Anti–Cancer Nano Drugs Delivery”, Am J Nanotechnol Nanomed. 1 (1): 001–009.
View at Publisher | View at Google Scholar - Heidari, (2018). “Vivo 1H or Proton NMR, 13C NMR, 15N NMR and 31P NMR Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Ann Biomet Biostat. 1 (1): 1001.
View at Publisher | View at Google Scholar - Heidari, (2018). “Grazing–Incidence Small–Angle Neutron Scattering (GISANS) and Grazing–Incidence X–Ray Diffraction (GIXD) Comparative Study on Malignant and Benign Human Cancer Cells, Tissues and Tumors under Synchrotron Radiation”, Ann Cardiovasc Surg. 1 (2): 1006.
View at Publisher | View at Google Scholar - Heidari, (2018). “Adsorption Isotherms and Kinetics of Multi–Walled Carbon Nanotubes (MWCNTs), Boron Nitride Nanotubes (BNNTs), Amorphous Boron Nitride Nanotubes (a–BNNTs) and Hexagonal Boron Nitride Nanotubes (h–BNNTs) for Eliminating Carcinoma, Sarcoma, Lymphoma, Leukemia, Germ Cell Tumor and Blastoma Cancer Cells and Tissues”, Clin Med Rev Case Rep 5: 201.
View at Publisher | View at Google Scholar - Heidari, (2018). “Correlation Spectroscopy (COSY), Exclusive Correlation Spectroscopy (ECOSY), Total Correlation Spectroscopy (TOCSY), Incredible Natural–Abundance Double–Quantum Transfer Experiment (INADEQUATE), Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC), Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC), Nuclear Overhauser Effect Spectroscopy (NOESY) and Rotating Frame Nuclear Overhauser Effect Spectroscopy (ROESY) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Acta Scientific Pharmaceutical Sciences 2.5: 30–35.
View at Publisher | View at Google Scholar - Heidari, (2018). “Small–Angle X–Ray Scattering (SAXS), Ultra–Small Angle X–Ray Scattering (USAXS), Fluctuation X–Ray Scattering (FXS), Wide–Angle X–Ray Scattering (WAXS), Grazing–Incidence Small–Angle X–Ray Scattering (GISAXS), Grazing–Incidence Wide–Angle X–Ray Scattering (GIWAXS), Small–Angle Neutron Scattering (SANS), Grazing–Incidence Small–Angle Neutron Scattering (GISANS), X–Ray Diffraction (XRD), Powder X–Ray Diffraction (PXRD), Wide–Angle X–Ray Diffraction (WAXD), Grazing–Incidence X–Ray Diffraction (GIXD) and Energy–Dispersive X–Ray Diffraction (EDXRD) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Oncol Res Rev, Volume 1 (1): 1–10.
View at Publisher | View at Google Scholar - Heidari, (2018). “Pump–Probe Spectroscopy and Transient Grating Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Adv Material Sci Engg, Volume 2, Issue 1, Pages 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “Grazing–Incidence Small–Angle X–Ray Scattering (GISAXS) and Grazing–Incidence Wide–Angle X–Ray Scattering (GIWAXS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Insights Pharmacol Pharm Sci 1 (1): 1–8.
View at Publisher | View at Google Scholar - Heidari, (2018). “Acoustic Spectroscopy, Acoustic Resonance Spectroscopy and Auger Spectroscopy Comparative Study on Anti–Cancer Nano Drugs Delivery in Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Nanosci Technol 5 (1): 1–9.
View at Publisher | View at Google Scholar - Heidari, (2018). “Niobium, Technetium, Ruthenium, Rhodium, Hafnium, Rhenium, Osmium and Iridium Ions Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Nanomed Nanotechnol, 3 (2): 000138.
View at Publisher | View at Google Scholar - Heidari, (2018). “Homonuclear Correlation Experiments Such as Homonuclear Single–Quantum Correlation Spectroscopy (HSQC), Homonuclear Multiple–Quantum Correlation Spectroscopy (HMQC) and Homonuclear Multiple–Bond Correlation Spectroscopy (HMBC) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Austin J Proteomics Bioinform & Genomics. 5 (1): 1024.
View at Publisher | View at Google Scholar - Heidari, (2018). “Atomic Force Microscopy Based Infrared (AFM–IR) Spectroscopy and Nuclear Resonance Vibrational Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, J Appl Biotechnol Bioeng. 5 (3): 142‒148.
View at Publisher | View at Google Scholar - Heidari, (2018). “Time–Dependent Vibrational Spectral Analysis of Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, J Cancer Oncol, 2 (2): 000124.
View at Publisher | View at Google Scholar - Heidari, (2018). “Palauamine and Olympiadane Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Arc Org Inorg Chem Sci 3 (1).
View at Publisher | View at Google Scholar - R. Gobato, A. Heidari, (2018). “Infrared Spectrum and Sites of Action of Sanguinarine by Molecular Mechanics and Ab Initio Methods”, International Journal of Atmospheric and Oceanic Sciences. Vol. 2, No. 1, pp. 1–9.
View at Publisher | View at Google Scholar - Heidari, (2018). “Angelic Acid, Diabolic Acids, Draculin and Miraculin Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Med & Analy Chem Int J, 2 (1): 000111.
View at Publisher | View at Google Scholar - Heidari, (2018). “Gamma Linolenic Methyl Ester, 5–Heptadeca–5,8,11–Trienyl 1,3,4–Oxadiazole–2–Thiol, Sulphoquinovosyl Diacyl Glycerol, Ruscogenin, Nocturnoside B, Protodioscine B, Parquisoside–B, Leiocarposide, Narangenin, 7–Methoxy Hespertin, Lupeol, Rosemariquinone, Rosmanol and Rosemadiol Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Int J Pharma Anal Acta, 2 (1): 007–014.
View at Publisher | View at Google Scholar - Heidari, (2018). “Fourier Transform Infrared (FTIR) Spectroscopy, Attenuated Total Reflectance Fourier Transform Infrared (ATR–FTIR) Spectroscopy, Micro–Attenuated Total Reflectance Fourier Transform Infrared (Micro–ATR–FTIR) Spectroscopy, Macro–Attenuated Total Reflectance Fourier Transform Infrared (Macro–ATR–FTIR) Spectroscopy, Two–Dimensional Infrared Correlation Spectroscopy, Linear Two–Dimensional Infrared Spectroscopy, Non–Linear Two–Dimensional Infrared Spectroscopy, Atomic Force Microscopy Based Infrared (AFM–IR) Spectroscopy, Infrared Photodissociation Spectroscopy, Infrared Correlation Table Spectroscopy, Near–Infrared Spectroscopy (NIRS), Mid–Infrared Spectroscopy (MIRS), Nuclear Resonance Vibrational Spectroscopy, Thermal Infrared Spectroscopy and Photothermal Infrared Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, Glob Imaging Insights, Volume 3 (2): 1–14.
View at Publisher | View at Google Scholar - Heidari, (2018). “Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC) and Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC) Comparative Study on Malignant and Benign Human Cancer Cells, Tissues and Tumors under Synchrotron and Synchrocyclotron Radiations”, Chronicle of Medicine and Surgery 2.3: 144–156, 2018.
View at Publisher | View at Google Scholar - Heidari, (2018). “Tetrakis [3, 5–bis (Trifluoromethyl) Phenyl] Borate (BARF)–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules”, Medical Research and Clinical Case Reports 2.1: 113–126.
View at Publisher | View at Google Scholar - Heidari, (2018). “Sydnone, Münchnone, Montréalone, Mogone, Montelukast, Quebecol and Palau’amine–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules”, Sur Cas Stud Op Acc J. 1 (3).
View at Publisher | View at Google Scholar - Heidari, (2018). “Fornacite, Orotic Acid, Rhamnetin, Sodium Ethyl Xanthate (SEX) and Spermine (Spermidine or Polyamine) Nanomolecules Incorporation into the Nanopolymeric Matrix (NPM)”, International Journal of Biochemistry and Biomolecules, Vol. 4: Issue 1, Pages 1–19.
View at Publisher | View at Google Scholar - Heidari, R. Gobato, (2018). “Putrescine, Cadaverine, Spermine and Spermidine–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules”, Parana Journal of Science and Education (PJSE)–v.4, n.5, (1–14) July 1.
View at Publisher | View at Google Scholar - Heidari, (2018). “Cadaverine (1,5–Pentanediamine or Pentamethylenediamine), Diethyl Azodicarboxylate (DEAD or DEADCAT) and Putrescine (Tetramethylenediamine) Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Hiv and Sexual Health Open Access Open Journal. 1 (1): 4–11.
View at Publisher | View at Google Scholar - Heidari, (2018). “Improving the Performance of Nano–Endofullerenes in Polyaniline Nanostructure–Based Biosensors by Covering Californium Colloidal Nanoparticles with Multi–Walled Carbon Nanotubes”, Journal of Advances in Nanomaterials, Vol. 3, No. 1, Pages 1–28.
View at Publisher | View at Google Scholar - R. Gobato, A. Heidari, (2018). “Molecular Mechanics and Quantum Chemical Study on Sites of Action of Sanguinarine Using Vibrational Spectroscopy Based on Molecular Mechanics and Quantum Chemical Calculations”, Malaysian Journal of Chemistry, Vol. 20 (1), 1–23.
View at Publisher | View at Google Scholar - Heidari, (2018). “Vibrational Biospectroscopic Studies on Anti–Cancer Nanopharmaceuticals (Part I)”, Malaysian Journal of Chemistry, Vol. 20 (1), 33–73.
View at Publisher | View at Google Scholar - Heidari, (2018). “Vibrational Biospectroscopic Studies on Anti–Cancer Nanopharmaceuticals (Part II)”, Malaysian Journal of Chemistry, Vol. 20 (1), 74–117.
View at Publisher | View at Google Scholar - Heidari, (2018). “Uranocene (U(C8H8)2) and Bis(Cyclooctatetraene)Iron (Fe(C8H8)2 or Fe(COT)2)–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules”, Chemistry Reports, Vol. 1, Iss. 2, Pages 1–16.
View at Publisher | View at Google Scholar - Heidari, (2018). “Biomedical Systematic and Emerging Technological Study on Human Malignant and Benign Cancer Cells and Tissues Biospectroscopic Analysis under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (3): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “Deep–Level Transient Spectroscopy and X–Ray Photoelectron Spectroscopy (XPS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Res Dev Material Sci. 7(2). RDMS.000659.
View at Publisher | View at Google Scholar - Heidari, (2018). “C70–Carboxyfullerenes Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Glob Imaging Insights, Volume 3 (3): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “The Effect of Temperature on Cadmium Oxide (CdO) Nanoparticles Produced by Synchrotron Radiation in the Human Cancer Cells, Tissues and Tumors”, International Journal of Advanced Chemistry, 6 (2) 140–156.
View at Publisher | View at Google Scholar - Heidari, (2018). “A Clinical and Molecular Pathology Investigation of Correlation Spectroscopy (COSY), Exclusive Correlation Spectroscopy (ECOSY), Total Correlation Spectroscopy (TOCSY), Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC) and Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC) Comparative Study on Malignant and Benign Human Cancer Cells, Tissues and Tumors under Synchrotron and Synchrocyclotron Radiations Using Cyclotron versus Synchrotron, Synchrocyclotron and the Large Hadron Collider (LHC) for Delivery of Proton and Helium Ion (Charged Particle) Beams for Oncology Radiotherapy”, European Journal of Advances in Engineering and Technology, 5 (7): 414–426.
View at Publisher | View at Google Scholar - Heidari, (2018). “Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, J Oncol Res; 1 (1): 1–20.
View at Publisher | View at Google Scholar - Heidari, (2018). “Use of Molecular Enzymes in the Treatment of Chronic Disorders”, Canc Oncol Open Access J. 1 (1): 12–15.
View at Publisher | View at Google Scholar - Heidari, (2018). “Vibrational Biospectroscopic Study and Chemical Structure Analysis of Unsaturated Polyamides Nanoparticles as Anti–Cancer Polymeric Nanomedicines Using Synchrotron Radiation”, International Journal of Advanced Chemistry, 6 (2) 167–189.
View at Publisher | View at Google Scholar - Heidari, (2018). “Adamantane, Irene, Naftazone and Pyridine–Enhanced Precatalyst Preparation Stabilization and Initiation (PEPPSI) Nano Molecules”, Madridge J Nov Drug Res. 2 (1): 61–67.
View at Publisher | View at Google Scholar - Heidari, (2018). “Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC) and Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Madridge J Nov Drug Res, 2 (1): 68–74.
View at Publisher | View at Google Scholar - Heidari, R. Gobato, (2018). “A Novel Approach to Reduce Toxicities and to Improve Bioavailabilities of DNA/RNA of Human Cancer Cells–Containing Cocaine (Coke), Lysergide (Lysergic Acid Diethyl Amide or LSD), Δ⁹–Tetrahydrocannabinol (THC) [(–)–trans–Δ⁹–Tetrahydrocannabinol], Theobromine (Xantheose), Caffeine, Aspartame (APM) (NutraSweet) and Zidovudine (ZDV) [Azidothymidine (AZT)] as Anti–Cancer Nano Drugs by Coassembly of Dual Anti–Cancer Nano Drugs to Inhibit DNA/RNA of Human Cancer Cells Drug Resistance”, Parana Journal of Science and Education (PJSE), v. 4, n. 6, pp. 1–17.
View at Publisher | View at Google Scholar - Heidari, R. Gobato, (2018). “Ultraviolet Photoelectron Spectroscopy (UPS) and Ultraviolet–Visible (UV–Vis) Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Parana Journal of Science and Education (PJSE), v. 4, n. 6, pp. 18–33.
View at Publisher | View at Google Scholar - R. Gobato, A. Heidari, A. Mitra, (2018). “The Creation of C13H20BeLi2SeSi. The Proposal of a Bio–Inorganic Molecule, Using Ab Initio Methods for the Genesis of a Nano Membrane”, Arc Org Inorg Chem Sci 3 (4). AOICS.MS.ID.000167.
View at Publisher | View at Google Scholar - R. Gobato, A. Heidari, (2018). “Using the Quantum Chemistry for Genesis of a Nano Biomembrane with a Combination of the Elements Be, Li, Se, Si, C and H”, J Nanomed Res.7 (4): 241‒252.
View at Publisher | View at Google Scholar - Heidari, (2018). “Bastadins and Bastaranes–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules”, Glob Imaging Insights, Volume 3 (4): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “Fucitol, Pterodactyladiene, DEAD or DEADCAT (DiEthyl AzoDiCArboxylaTe), Skatole, the NanoPutians, Thebacon, Pikachurin, Tie Fighter, Spermidine and Mirasorvone Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Glob Imaging Insights, Volume 3 (4): 1–8.
View at Publisher | View at Google Scholar - E. Dadvar, A. Heidari, (2018). “A Review on Separation Techniques of Graphene Oxide (GO)/Base on Hybrid Polymer Membranes for Eradication of Dyes and Oil Compounds: Recent Progress in Graphene Oxide (GO)/Base on Polymer Membranes–Related Nanotechnologies”, Clin Med Rev Case Rep 5: 228.
View at Publisher | View at Google Scholar - Heidari, R. Gobato, (2018). “First–Time Simulation of Deoxyuridine Monophosphate (dUMP) (Deoxyuridylic Acid or Deoxyuridylate) and Vomitoxin (Deoxynivalenol (DON)) ((3α,7α)–3,7,15–Trihydroxy–12,13–Epoxytrichothec–9–En–8–One)–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Parana Journal of Science and Education (PJSE), Vol. 4, No. 6, pp. 46–67.
View at Publisher | View at Google Scholar - Heidari, (2018). “Buckminsterfullerene (Fullerene), Bullvalene, Dickite and Josiphos Ligands Nano Molecules Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Hematology and Thromboembolic Diseases Prevention, Diagnosis and Treatment under Synchrotron and Synchrocyclotron Radiations”, Glob Imaging Insights, Volume 3 (4): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “Fluctuation X–Ray Scattering (FXS) and Wide–Angle X–Ray Scattering (WAXS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (4): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “A Novel Approach to Correlation Spectroscopy (COSY), Exclusive Correlation Spectroscopy (ECOSY), Total Correlation Spectroscopy (TOCSY), Incredible Natural–Abundance Double–Quantum Transfer Experiment (INADEQUATE), Heteronuclear Single–Quantum Correlation Spectroscopy (HSQC), Heteronuclear Multiple–Bond Correlation Spectroscopy (HMBC), Nuclear Overhauser Effect Spectroscopy (NOESY) and Rotating Frame Nuclear Overhauser Effect Spectroscopy (ROESY) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (5): 1–9.
View at Publisher | View at Google Scholar - Heidari, (2018). “Terphenyl–Based Reversible Receptor with Rhodamine, Rhodamine–Based Molecular Probe, Rhodamine–Based Using the Spirolactam Ring Opening, Rhodamine B with Ferrocene Substituent, Calix[4]Arene–Based Receptor, Thioether + Aniline–Derived Ligand Framework Linked to a Fluorescein Platform, Mercuryfluor–1 (Flourescent Probe), N,N’–Dibenzyl–1,4,10,13–Tetraraoxa–7,16–Diazacyclooctadecane and Terphenyl–Based Reversible Receptor with Pyrene and Quinoline as the Fluorophores–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules”, Glob Imaging Insights, Volume 3 (5): 1–9.
View at Publisher | View at Google Scholar - Heidari, (2018). “Small–Angle X–Ray Scattering (SAXS), Ultra–Small Angle X–Ray Scattering (USAXS), Fluctuation X–Ray Scattering (FXS), Wide–Angle X–Ray Scattering (WAXS), Grazing–Incidence Small–Angle X–Ray Scattering (GISAXS), Grazing–Incidence Wide–Angle X–Ray Scattering (GIWAXS), Small–Angle Neutron Scattering (SANS), Grazing–Incidence Small–Angle Neutron Scattering (GISANS), X–Ray Diffraction (XRD), Powder X–Ray Diffraction (PXRD), Wide–Angle X–Ray Diffraction (WAXD), Grazing– Incidence X–Ray Diffraction (GIXD) and Energy–Dispersive X–Ray Diffraction (EDXRD) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (5): 1–10.
View at Publisher | View at Google Scholar - Heidari, (2018). “Nuclear Resonant Inelastic X–Ray Scattering Spectroscopy (NRIXSS) and Nuclear Resonance Vibrational Spectroscopy (NRVS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (5): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “Small–Angle X–Ray Scattering (SAXS) and Ultra–Small Angle X–Ray Scattering (USAXS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (5): 1–7.
View at Publisher | View at Google Scholar - Heidari, (2018). “Curious Chloride (CmCl3) and Titanic Chloride (TiCl4)–Enhanced Precatalyst Preparation Stabilization and Initiation (EPPSI) Nano Molecules for Cancer Treatment and Cellular Therapeutics”, J. Cancer Research and Therapeutic Interventions, Volume 1, Issue 1, Pages 01–10.
View at Publisher | View at Google Scholar - R. Gobato, M. R. R. Gobato, A. Heidari, A. Mitra, (2018). “Spectroscopy and Dipole Moment of the Molecule C13H20BeLi2SeSi via Quantum Chemistry Using Ab Initio, Hartree–Fock Method in the Base Set CC–pVTZ and 6–311G**(3df, 3pd)”, Arc Org Inorg Chem Sci 3 (5), Pages 402–409.
View at Publisher | View at Google Scholar - Heidari, (2018). “C60 and C70–Encapsulating Carbon Nanotubes Incorporation into the Nano Polymeric Matrix (NPM) by Immersion of the Nano Polymeric Modified Electrode (NPME) as Molecular Enzymes and Drug Targets for Human Cancer Cells, Tissues and Tumors Treatment under Synchrotron and Synchrocyclotron Radiations”, Integr Mol Med, Volume 5 (3): 1–8.
View at Publisher | View at Google Scholar - Heidari, (2018). “Two–Dimensional (2D) 1H or Proton NMR, 13C NMR, 15N NMR and 31P NMR Spectroscopy Comparative Study on Malignant and Benign Human Cancer Cells and Tissues under Synchrotron Radiation with the Passage of Time”, Glob Imaging Insights, Volume 3 (6): 1–8.
View at Publisher | View at Google Scholar - Heidari, (2018). “FT–Raman Spectroscopy, Coherent Anti–Stokes Raman Spectroscopy (CARS) and Raman Optical Activity Spectroscopy (ROAS) Comparative Study on Malignant and Benign Human Cancer Cells and Tissues with the Passage of Time under Synchrotron Radiation”, Glob Imaging Insights, Volume 3 (6): 1–8.
View at Publisher | View at Google Scholar - Heidari, (2018). “A Modern and Comprehensive Investigation of Inelastic Electron Tunneling Spectroscopy (IETS) and Scanning Tunneling Spectroscopy on Malignant and Benign Human Cancer Cells, Tissues and Tumors through Optimizing Synchrotron Microbeam Radiotherapy for Human Cancer Treatments and Diagnostics: An Experimental Biospectroscopic Comparative Study”, Glob Imaging Insights, Volume 3 (6): 1–8.
View at Publisher | View at Google Scholar

 Clinic
Clinic
